# Install if not already available
# install.packages('ecmwfr')
# install.packages('terra')
# install.packages('tidyverse')
library(ecmwfr)
library(terra)
library(tidyverse)Downloading and Processing CERRA Climate Data from CDS
Introduction
This document outlines a complete workflow to download and process CERRA reanalysis climate data from the Copernicus Climate Data Store (CDS). The workflow uses the ecmwfr package to request data and terra to efficiently load and manipulate large gridded datasets. It covers:
- Authentication with the CDS API
- Request construction for multi-year climate data
- Batch data download
- Data loading and memory optimization
- Subsetting and aggregating the raster time series
1. Load Required Packages
We use two primary packages:
ecmwfr: for accessing and downloading data from CDSterra: for handling and analyzing large gridded datasets
2. Explore Available Datasets
We fetch the list of available datasets and filter those related to CERRA.
# Retrieve dataset catalog
cds_datasets <- ecmwfr::wf_datasets(service = 'cds')
# Filter for CERRA datasets
cerra_datasets <- cds_datasets[grepl('cerra', cds_datasets$name), ]3. Set CDS API Key
To download data from CDS, create an account at
👉 cds.climate.copernicus.eu
Then:
- Find your API Key in your profile
- Use the
wf_set_key()function once to store the credentials
# Replace with your own credentials
API_KEY <- '' # Your API key from CDS profile
USER <- '' # Your CDS username/email
# Set API key (only needs to be done once per machine)
wf_set_key(key = API_KEY, user = USER)4. Set CDS API Key
Before download, make sure that you signed the terms of use (see bottom of dataset page)
👉 CERRA terms of use
5. Define Download Parameters
Specify the years, months, days, times, and variable for which to download data. To understand the arguments please navigate to the dataset download page, play with the selection of the variables and check the API call at the bottom of the page.
# Define time dimensions and variable
dataset <- "reanalysis-cerra-single-levels"
years <- c(1984:2024)
months <- sprintf("%02d", 1:12)
days <- sprintf("%02d", 1:31)
times <- sprintf("%02d:00", seq(0, 21, by = 3))[1]
leadtime_hour <- 24
variable <- "total_precipitation"
# Output directory for downloaded files
path <- '/Volumes/Extreme SSD/TEMP/'6. Create a Request List
Generate a list of download requests, one for each year.
# Create list of requests (one per year)
request_list <- lapply(years, function(x)
list(
dataset_short_name = dataset,
data_type = "reanalysis",
level_type = "surface_or_atmosphere",
product_type = "forecast",
variable = variable,
year = x,
month = months,
day = days,
time = times,
leadtime_hour = leadtime_hour,
data_format = "grib",
target = paste0(variable, '_', x, ".grib")
)
)7. Send Download Requests
You can run a single request (for testing) or batch multiple years in parallel.
# Single request example
test <- ecmwfr::wf_request(
request = request_list[[1]],
path = path,
user = USER # see above
)
# Batch request (first two years as an example)
SET <- request_list[1:2]
test <- ecmwfr::wf_request_batch(
request_list = SET,
workers = length(SET), # workers set to the number of files
user = USER, # see above
path = path
)8. Load and Process the Downloaded Data
We use terra to efficiently load and work with the GRIB files.
a. Optimize Memory Usage
# Set up a temporary directory for terra to store intermediate files
tempdir <- tempfile(pattern = paste0("GLOB"), tmpdir = path)
dir.create(tempdir)
terra::terraOptions(tempdir = tempdir, verbose = TRUE)
# Clear memory
gc() used (Mb) gc trigger (Mb) limit (Mb) max used (Mb)
Ncells 1670259 89.3 3115932 166.5 NA 2444515 130.6
Vcells 2390926 18.3 8388608 64.0 102400 3264307 25.0b. Load Data Files
# List GRIB files
files <- list.files(pattern = variable, path = path, full.names = TRUE)
# Load the raster stack
data <- terra::rast(files)
# Extract time information and assign as layer names
ts <- terra::time(data)
# rename the raster list to timestamps
names(data) <- ts9. Subset the Raster Time Series
You can for instance subset by:
- Specific datetime
- Specific date
- Specific time
# Subset by exact datetime
subset <- data[[grep("1985-01-01", ts, fixed = TRUE)]]
terra::plot(subset)
gc()
# Subset by month
subset <- data[[grep("1985-01-", ts, fixed = TRUE)]]
terra::plot(subset)
gc()10. Area selection
To extract values from a raster within a specific region, combine crop() and mask().
- crop() limits the raster to the bounding box of a polygon (faster, rough cut).
- mask() sets cells outside the actual shape of the polygon to NA (precise shape).
# Luxembourg provinces
v <- terra::vect(system.file("ex/lux.shp", package = "terra"))
# Change projection system of the vector to match CERRA
v <- terra::project(v,data)
# Crop and mask
subset <- mask(crop(data, v), v)
gc()
# Calculate summary stats for each polygon
global_stats <- terra::global(subset, fun = c("min", "max", "mean", "sum", "range", "rms", "sd", "std"), na.rm = TRUE)
global_median <- terra::global(subset, fun = median, na.rm = TRUE)
# Calculate summary stats per pixel across the time series
min_r <- min(subset, na.rm = TRUE)
max_r <- max(subset, na.rm = TRUE)
mean_r <- mean(subset, na.rm = TRUE)
sum_r <- sum(subset, na.rm = TRUE)
range_r <- max(subset, na.rm = TRUE) - min(subset, na.rm = TRUE)
sd_r <- stdev(subset, na.rm = TRUE)
std_r <- stdev(subset, na.rm = TRUE,pop=TRUE)
median_r <- median(subset,na.rm = TRUE)
quantile_r <- app(subset,quantile,na.rm=TRUE) # quantiles
decile_r <- app(subset,quantile,probs=seq(0, 1, 0.1),na.rm=TRUE) # deciles 11. Monthly summary
a. Rasters
Here we compute monthly European level rasters giving the total precipitation per day.
ts <- terra::time(data) # a POSIXct vector, one for each layer
months <- substr(ts,1,7) # convert to date only (drop time part)
unique_months <- unique(months) # Unique dates in your dataset
# calculate the daily accumulated rainfall per pixel across Europe
monthly_sum <- lapply(unique_months, function(d) {
idx <- which(months == d) # layers for this day
subset_day <- data[[idx]] # subset raster layers
sum_day <- sum(subset_day, na.rm = TRUE) # per-pixel mean
names(sum_day) <- as.character(d) # name the layer
gc() # free up memory
return(sum_day)
})
# convert list to spatraster
monthly_sum <- terra::rast(monthly_sum)
# plot first 10 days of total precipitation
plot(monthly_sum[[1:12]])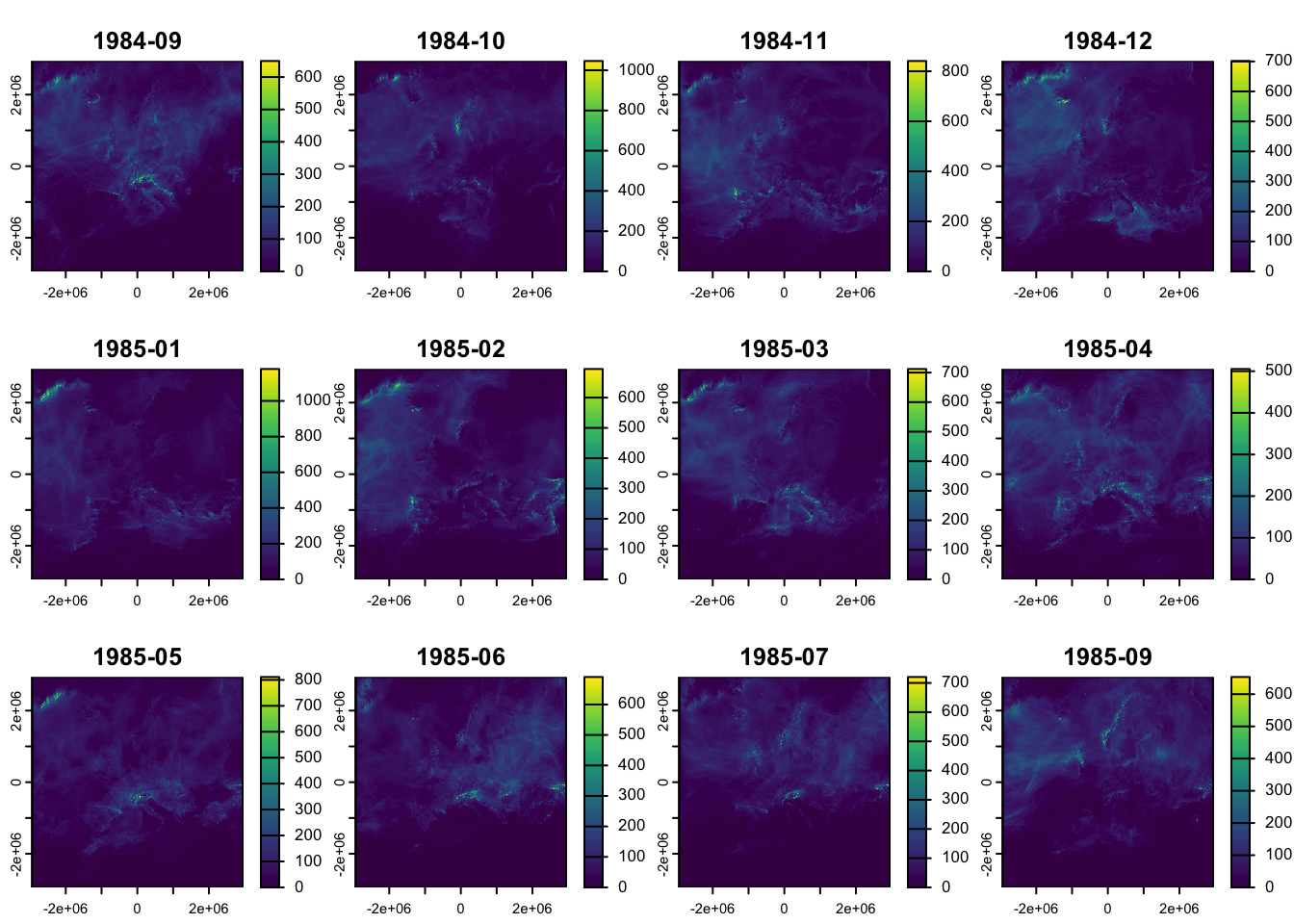
# visualize total precipitation on Sept 5th 1984 on a zoomable map
plet(monthly_sum[[5]])b. Extract values for point locations
Here we extract the daily total precipitation for 12 centroids of Luxembourg provinces.
# extract daily accumulated rainfall for the centroids (point location) of each province in Luxembourg
v <- vect(system.file("ex/lux.shp", package = "terra"))
v <- project(v,monthly_sum)
# convert polygons to points using their centroids
p <- terra::centroids(v)
# extract values from the above daily rasters
values <- terra::extract(monthly_sum,p,xy=TRUE)
# add province names
values$province <- p$NAME_2
# Pivot longer: gather date columns into one
monthly_rainfall <- values %>%
pivot_longer(
cols = -c(ID, x, y, province),
names_to = "month",
values_to = "value"
) %>%
mutate(month = as.Date(paste0(month,'-01')))
# daily rainfall per point
ggplot(monthly_rainfall, aes(x = month, y = value)) +
geom_line(color = "darkgreen") +
facet_wrap(~ province) +
labs(title = "Daily Rainfall at Centroids of the provinces of Luxembourg",
x = "Date", y = "Rainfall (mm)") +
theme_minimal()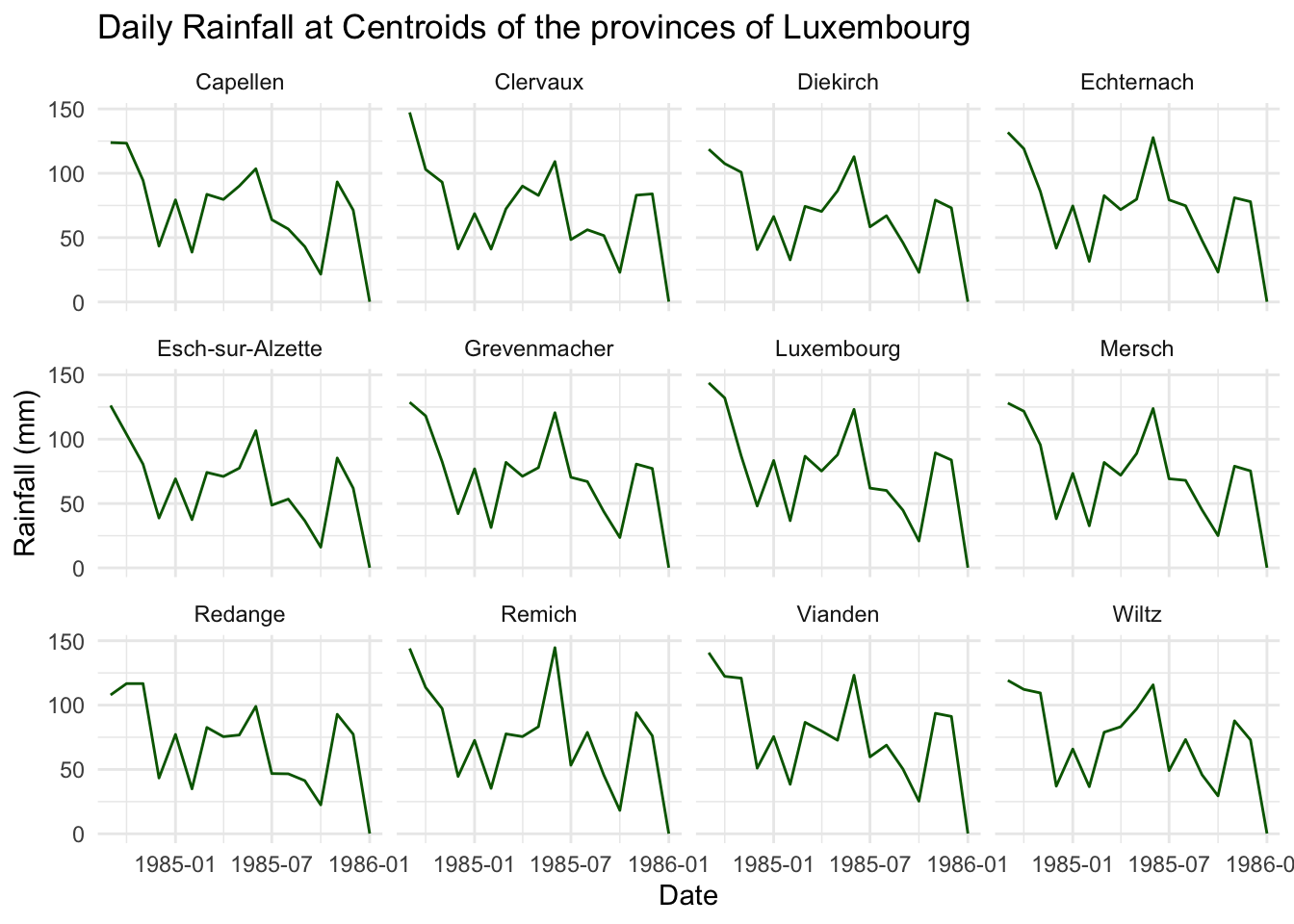
Conclusion
This script provides a complete automated workflow for downloading and processing total precipitation data from the CERRA dataset using CDS. It supports batch downloading, memory-optimized data handling, and time-based subsetting, making it suitable for long-term climatological analysis or integration into larger pipelines.
Downloading ECMWF datasets via Python
Some ECMWF datasets are not accessible through the R package. For instance, datasets in the xds (developmental) platform. For those datasets python can be used.
Set up python environment
# Create environment
conda create -n drought_env python=3.11 -y
# Activate it
conda activate drought_env
# Install required packages
pip install cdsapi tqdmSet the xds credentials
See profile on the xds website for your credentials.
# create the file
nano ~/.cdsapirc
# configure
url: https://xds-preprod.ecmwf.int/api
key: API-KEYCreate a python script
# create file with nano
nano download_drought_data.py# paste script
import cdsapi
from tqdm import tqdm
import os
import sys
# years as arguments
if len(sys.argv) < 2:
print("Usage: python download_drought_data.py <year1> <year2> ...")
print("Example: python download_drought_data.py 1941 1942 1943")
sys.exit(1)
years = [str(y) for y in sys.argv[1:]] # list of years from CLI args
# Initialize CDS client
c = cdsapi.Client()
dataset = "derived-drought-historical-monthly"
base_request = {
"variable": [
"standardised_precipitation_evapotranspiration_index"
],
"accumulation_period": ["24"],
"version": "1_0",
"product_type": ["reanalysis"],
"dataset_type": "consolidated_dataset",
"month": [
"01", "02", "03",
"04", "05", "06",
"07", "08", "09",
"10", "11", "12"
]
}
# Output directory
outdir = "./downloads"
os.makedirs(outdir, exist_ok=True)
# Years to download
years = [str(y) for y in range(1941, 2025)]
# Loop over years
for year in tqdm(years, desc="Downloading drought data"):
target = os.path.join(outdir, f"drought_{year}")
if os.path.exists(target):
print(f"Skipping {year} (already exists).")
continue
request = base_request.copy()
request["year"] = [year]
try:
c.retrieve(dataset, request, target)
except Exception as e:
print(f"Failed to download {year}: {e}")run python download script
Run the python script to download the datasets and extract the zipped files.
# run python script to download the data as zips
python download_drought_data.py $(seq 1941 2025)
# extract downloaded zips
cd downloads
for f in drought_*; do
echo "Extracting $f ..."
unzip -o "$f"
doneOnce unzipped you can list all .nc files and import them in R with terra::rast().
filenames <- list.files(pattern='.nc',recursive = T,full.names = T)
r <- terra::rast(filenames)